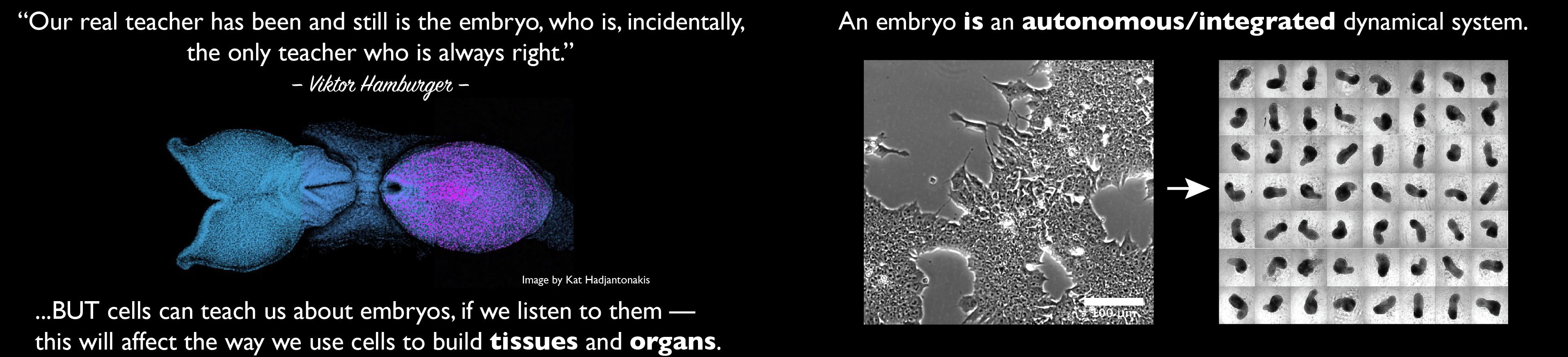
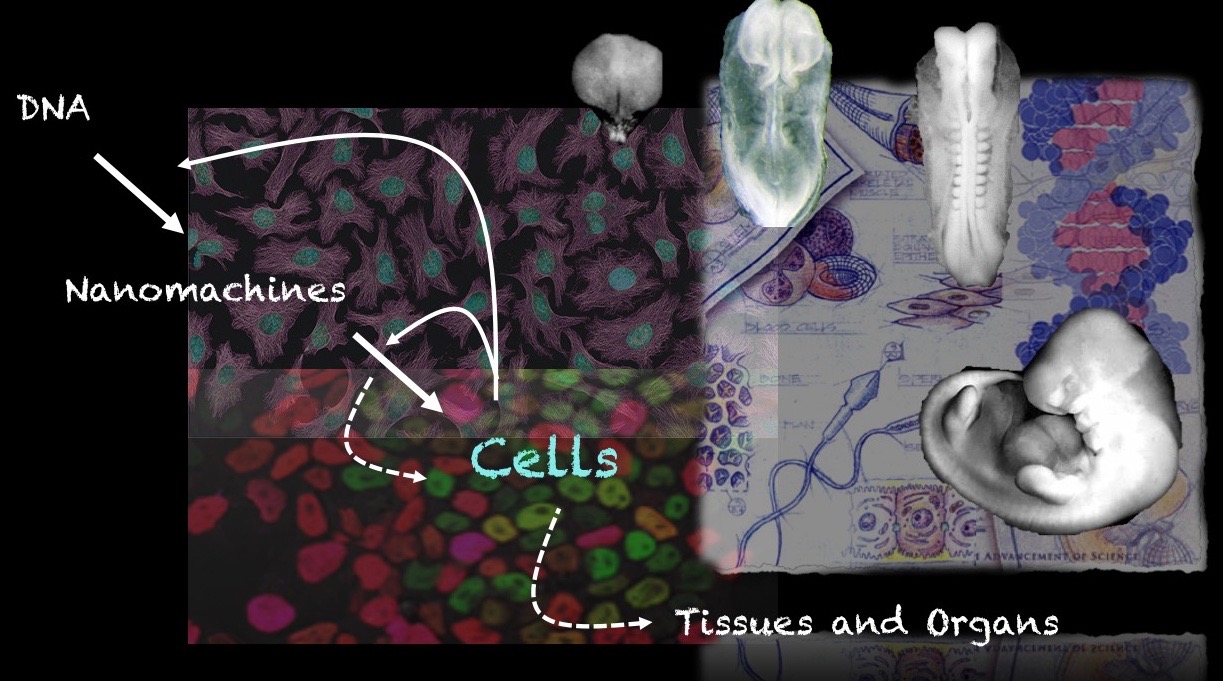
Our group is interested in how cells build embryos. We approach this question by asking cells, specifically Embryonic Stem Cells (ESCs) how much can they do on their own. We work with mammalian ESCs because over the last decade advances in our ability to grow them and differentiate them in dishes and wells have opened up exciting experimental possibilities that we are exploiting. In these endeavours, we don’t forget the embryo which is, perhaps not the teacher that Viktor Hamburger anointed it as in his famous sentence, but a reference for our work.
A few years ago, we saw that under defined culture conditions mouse ESCs could be coaxed into 3D aggregates that would form structures with a good representation of the vertebrate body plan. We called these structures ‘gastruloids’ because what they create mirrors the outcome of gastrulation.
You can find more information about gastruloids and what we are doing with them in these pages.
Latest Posts
These are difficult times. COVID-19 is changing the way we live and, for now — and maybe for the future — the way we work. The loss of life and the effect on the economy and society is a matter for reflection. Importantly, this disease has highlighted the central role that Science plays in our Society and the need to support and expand its base because Science does two things, deals with uncertainty and prepares for the unexpected by exploring what might not seem urgent today but will be key for the future. Furthermore, the enormous international efforts being done at the moment also highlight that Science is global and this, we suspect, will remain as such.
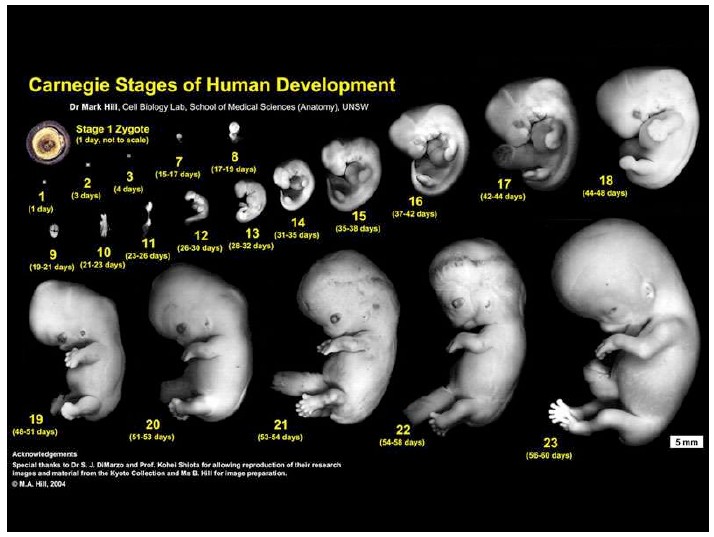
Notes on Human Embryos
III. Models of human development Pluripotent stem cells (PSCs: ESCs and iPSCs) create opportunities to address the ethical and technical constraints associated with the use of human embryos. In order to discuss and evaluate these opportunities it is important first to provide a context for this work and look at its roots and assess its limitations. These days one has the impression that Embryonic Stem Cells (ESCs) are collections of genes, of epigenetic modifications all revealed by barcodes and sophisticated […]
Recent Publications
Moris, N., Anlas, K., van den Brink, S.C., et al. An in vitro model of early anteroposterior organization during human development. Nature (2020).
van den Brink, S.C., Alemany, A., van Batenburg, V. et al. Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature (2020).
Beccari, L., Moris, N., Girgin, M. et al. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature (2018).
Collaborators
Dr Ben Steventon (Department of Genetics, University of Cambridge, UK).
Professor Jordi Garcia Ojalvo (DCEXS, Universitat Pompeu Fabra, Barcelona, Spain)
Professor Alexander van Oudenaarden (Hubrecht Institute, Utrecht, The Netherlands).
Professor Matthias Lütolf (EPFL, Lausanne, Switzerland).
Professor Kat Hadjantonakis (Sloan Kettering, New York, USA).